Journal of Animal Research: v.10 n.1, p. 133-136. February 2020
DOI: 10.30954/2277-940X.01.2020.19
IGF-1 Gene Polymorphism in Migratory Gaddi Goats of Western Himalayan Region, India
ABSTRACT
Goat production is a predominant livestock activity in harsh climatic regions of the country particularly hilly regions. ‘Gaddi’ is a prominent goat breed of Himachal Pradesh constituting 60-65% of total goat population of 11.20 lakh (19th Livestock Census, 2012). The somatotropic axis has a key role in postnatal growth and metabolism. IGF-1 gene encodes the protein that is structurally and functionally similar to insulin and regulates cellular synthesis of DNA as well as cellular growth and development, especially in neurons and also mediates the effect of GHgene. The present study was carried on‘Gaddi’, a distinct goat breed of high altitude areas of Western Himalayan region, for molecular characterization of IGF-1 gene and further analyse its association with growth traits. Blood samples from 63 genetically unrelated animals of Gaddi goat breed were taken from the migratory flocks under AICRP and Gaddi goat unit of CSKHPKV, Palampur and subjected to DNA isolation. 363 bp amplicon was generated and PCR-RFLP analysis using Hae III restriction enzyme (RE) revealed two variants (AB and BB) however, no significant association could be established. Allele frequencies for A and B alleles were 0.25 and 0.75. The estimates obtained for Ne, Hobs, Hexp and PIC were 1.61, 0.51, 0.38 and 0.31, respectively. PIC value of 0.31 implies the effectiveness of the marker in population studies and also revealed median level of polymorphism. Sequencing confirmed one nucleotide mutation (C264G), however, no significant association were found at IGF-1 genotype with biometric traits in screened Gaddi goats.
Keywords: IGF-1 gene, Genetic polymorphism, Gaddi goat, Migratory pastoralism
Goat production is a predominant livestock activity in harsh climatic regions of the country particularly in hilly areas and arid zones with little arable agricultural land. Gaddi, also known as White Himalayan goat is a prominent goat breed of mid to high hilly regions of the state of H.P. constituting 60-65% of total goat population of 11.20 lakh (19th Livestock Census, 2012) with its true home tract in H.P. but distribution extending to adjoining hilly areas of Jammu – Kashmir and Uttrakhand. It is mainly a meat breed with sturdy built and highly suitable to migratory production system. It is traditionally reared by nomadic “Gaddi” shepherds throughout the hilly region of the state and is considered the best suited goat breed for the prevalent migratory sheep and goat production system. Rapid growth as reflected by faster body weight gain, is important in meat goats as it influences net economic returns from sale of animals. Physiological regulation of body growth in animals is under the control of several genes. The growth hormone gene and its receptors like IGF-1 have been identified as major genes in regulating growth. IGF-1, encoded by IGF-1 gene plays an important role in growth and has anabolic effects. The IGF-I gene has been considered to be a candidate marker associated with growth traits in livestock animals (Thue and Buchanan, 2002; Zhang et al, 2008; Wu-Jun et al,2010).Consequently, selection based on high levels of endogenous IGF-1 helps to explore and exploit superior growth potential. Polymorphism in such genes may have association with certain important growth traits and thus, can serve as useful markers in marker assisted selection (MAS) for higher growth.
How to cite this article: Gitanjali, Sankhyan, V., Thakur, Y.P. and P.K. Dogra. (2020). IGF-1 gene polymorphism in migratory Gaddi goats of western Himalayan region, India. J. Anim. Res., 10(1): 133-136.
To analyze polymorphism of IGF-1 gene in Gaddi goats, PCR-RFLP technique was used. Samples were collected from 63 unrelated animals of Gaddi goats. Genomic DNA was isolated by phenol chloroform extraction technique (Sambrook and Russsel, 2001).
MATERIAL AND METHODS
The experimental animals for the present investigation were taken from adopted migratory flocks under AICRP on goat improvement and Gaddi goat unit of CSKHPKV, Palampur. Blood samples were collected from 63 genetically unrelated, randomly selected animals from the breeding tract as per the guidelines of FAO (1995) for measurement of domestic animal diversity (MoDAD). The information on growth records (body weight, body length, body height and heart girth) at different ages (3, 6, 9 and 12 months) for each animal that was considered for association studies was collected. 10 ml of blood was collected from the jugular vein of animals of different sex and age groups in sterile vacutainers containing anticoagulant. After collection, the tubes were transported to the laboratory in an icebox and stored at -20°C. Genomic DNA was isolated by utilizing phenol chloroform extraction technique, as described by Sambrook and Russel (2001). IGF-lgene was amplified using the forward primer (5’CACAGCGTATTATCCCAC3’) and reverse primer (5’ GACACTATGAGCCAGAAG3’), respectively (Liu et al,2012). 25 μl of PCR mixture was prepared by adding 10 pM of each primer, 10mM of dNTP mix, 2.5 mM MgCl2, 5X PCR buffer, 100 ng template DNA and 0.5 units of TaqDNA polymerase. The amplification was carried out with initial denaturation of 5 minutes at 94°C followed by 35 cycles of denaturation at 94°C for 1 minute, annealing at 56°C for 2 minute and extension at 72°C for 1 minute and lastly the final extension of 5 minute at 72°C. An aliquot of 15 pl of PCR product was digested overnight with 1unit of HaeIII RE at 37°C. Representative samples of different genotypes as revealed by PCR-RFLP were got sequenced after purification of respective PCR product.
Association studies
The following fixed effects model was used for analysis of growth traits in Gaddi goats and least square means were used for multiple comparison of body weights and body measurements for different genotypes.
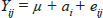
Yij = Observations for phenotypic value of growth characteristics (body weight, body length, heart girth and body height)
μ = overall mean of the trait
a. = fixed effect of genotypes (i = AA, AB and BB)
Analysis was done using the general linear model procedure of SAS (ver 9.3) (SAS Institute Inc, Cary, NC, USA). Mean separation procedures were conducted using a least significant difference test.
RESULTS AND DISCUSSION
PCR amplification of IGF-1 yielded 363 bp amplicon as depicted by Fig. 1. PCR-RFLP analysis of amplified products with HaeIII RE exhibited three fragments of 363 bp, 264 bops and 99 bp (Fig. 2) that revealed two genotypes viz., AB and BB. The allele frequencies for A and B alleles were estimated as 0.25 and 0.75, respectively. The frequency of AB and BB genotypes were 0.51 and 0.49.
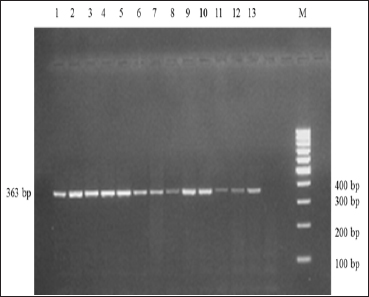
Fig. 1: Electrophoretic profile of amplified IGF-1 in 2% agarose gel. Lanes 1-13 represents amplicon size of 363 bp. M represents 100 bp DNA marker
Similarly, Deng et al. (2010) observed genotype frequencies of AG and GG for P1 locus as 0.15 and 0.85 in Guanzhong goats and 0.21 and 0.79 in Xinong Saanen goats. For P4 locus, genotype frequencies for CC, CG and GG genotypes were 0.10, 0.16 and 0.74 in Guanzhong breed and 0.08, 0.43 and 0.49 in Xinong Saanen breed. In another study of IGF-I gene in Cashmere goats, Liu et al. (2012) identified two alleles A and B with frequencies 0.75 and 0.25 and three genotypes AA (0.58), AB (0.33) and BB(0.09), respectively. Sankhyan et al. (2019) also reported similar findings in different sheep and goat breeds of Western Himalayan region with frequencies of A allele as 0.66, 0.59, 0.65 and 0.70 and B allele as 0.34, 0.41, 0.35 and 0.20 in Gaddi sheep, Rampur Bushair sheep, Gaddi goat and Chegu goat breeds.
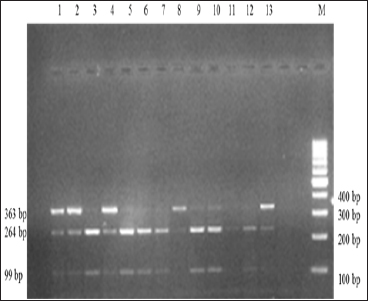
Fig. 2: Electrophoretic pattern obtained after HaeIII digestion of IGF-1 gene. Lanes 1,2,4 and 8-13 represented AB genotype. Lanes 3 and 5-7 represented BB genotype. M represented 100 bp DNA marker
Genetic diversity analysis and test for Hardy-Weinberg Equilibrium (HWE)
The IGF-1 gene showed polymorphism in the studied population of Gaddi goat breed. The measures of genetic diversity indicated considerable variation at the studied locus. The estimates obtained for Ne, Hobs, Hexp and PIC were 1.61, 0.51, 0.38 and 0.31, respectively. PlC value of 0.31 implies the effectiveness of the marker in population studies and also revealed median level of polymorphism in investigated Gaddi goat population. The absence of heterozygous deficiency at studied locus for Gaddi goats is indicated by negative FIS measure of -0.34. Wu-Jun et al. (2010) reported Hobs, Hexp and PIC values of 0.52, 0.47 and 0.36 in Chinese goats which were almost similar with the present study. The test for HWE indicated that Gaddi goat population was deviating significantly from equilibrium (P˂0.01). Wu-Jun et al. (2010) in Chinese goats and Negahdary et al. (2013) in Makooei sheep breed of Iran reported genotypic distribution at IGF-1 loci that were in agreement with HWE as seen in the present study.
Association of IGF-1 polymorphism with growth traits in Gaddi goat breed
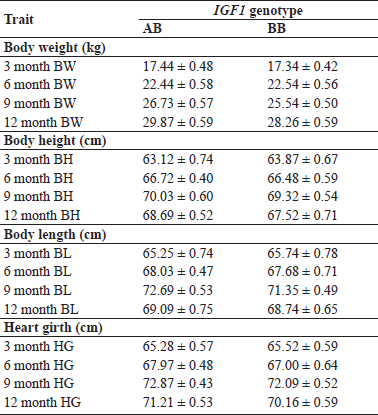
The IGF-1 genotypes of 63 animals belonging to Gaddi goat breed were screened along with the data on growth and body measurements traits (BW, BL, BH and HG) at different ages (3M, 6M, 9M, 12M). The association of IGF-1 variants with growth traits is depicted in Table 1. The mean body weights (kg) at 3M, 6M, 9M and 12M were estimated as 17.34 ± 0.42 and 17.44 ± 0.48, 22.44 ± 0.58 and 22.54 ± 0.56, 25.54 ± 0.50 and 26.73 ± 0.57 and 28.26 ± 0.59 and 29.87 ± 0.59, respectively for the detected AB and BB genotypes. The corresponding body heights (cm) at 3M, 6M, 9M and 12M were 63.12 ± 0.74 and 63.87 ± 0.67, 66.48 ± 0.59 and 66.72 ± 0.40, 69.32 ± 0.54 and 70.03 ± 0.60 and 67.52 ± 0.71 and 68.69 ± 0.52, respectively. The body lengths (cm) at 3M, 6M, 9M and 12M were 65.25 ± 0.74 and 65.74 ± 0.78, 67.68 ± 0.71 and 68.03 ± 0.47, 71.35 ± 0.49 and 72.69 ± 0.53 and 68.74 ± 0.65 and 69.09 ± 0.75, respectively for different genotypes. The heart girth (cm) was 65.28 ± 0.57 and 65.52 ± 0.59, 67.00 ± 0.64 and 67.97 ± 0.48, 72.09 ± 0.52 and 72.87 ± 0.43 and 70.16 ± 0.59 and 71.21 ± 0.53 for 3M, 6M, 9M and 12M.
No significant association could be observed between different variants of IGF-1 gene and growth traits in Gaddi goats. The earlier studies carried on various breeds also reported that the results differ in association between IGF- 1 polymorphism and growth traits. The findings of the present study were similar to those reported by Wang et al. (2011) with no association between birth weight and IGF- 1 polymorphism in three indigenous Chinese goat breeds. Similarly, Gholibeikifard et al. (2013) did not observe the effect of SNPs at exon-3 of IGF-1 gene on growth traits in Baluchi sheep. The results of present investigation were in agreement with those reported by Rasouli et al. (2017) in Markhoz goats with no significant effect of IGF- 1 polymorphism on birth weight and body weight at 6, 9 and 12 months of age.
Table 1: Mean body weights and body measurements at different ages depicting association with observed IGF-1 genotypes in Gaddi goats
CONCLUSION
In the present study, PCR-RFLP assay revealed polymorphism in the amplified product of IGF gene in migratory Gaddi goat population. DNA sequencing confirmed one nucleotide mutation (C264G) with allelic frequency of alleles A and B as 0.25 and 0.75, respectively. However, in present study no significant association were found at IGF-1 genotype with biometric traits in screened Gaddi goats.. Therefore, the study on additional data based on more number of animals in diversified flock should be carried out for further association studies.
Acknowledgements
Authors sincerely acknowledge the financial support received from Indian Council of Agricultural ResearchCentral Institute of Research on Goats (ICAR-CIRG), Makhdoom through All India Coordinated Research Project on Goat Improvement. Authors are also grateful for adopted flock owners for their cooperation.
Conflict of Interest: Authors declares that they do not have any conflict of interest.
REFERENCES
Deng, C.J., Ma, R.N., Yue, X.P., Lan, X.Y., Chen, H. and Lei C.Z. 2010. Association of IGF-I gene polymorphisms with milk yield and body size in Chinese dairy goats. Genet. Mol. Biol, 33: 266-270.
Gholibeikifard, A., Aminafshar, M. and Hosseinpour, M.M. 2013. Polymorphism of IGF-I and ADRB3 genes and their association with growth traits in the Iranian Baluchi sheep. J. Agr. Sci. Tech, 15: 1153-1162.
Liu, H., Liu, C., Yang, G., Li, H., Dai, J., Cong, Y. and Li, X. 2012. DNA polymorphism of IGFBP3 gene and its association with cashmere traits in Cashmere goats. Asian Austral. J. Anim. Sci, 25(11): 1515-1520.
Livestock Census. 2012. 19th Livestock Census, All India Report. Ministry of Agriculture Department of Animal Husbandry, Dairying and Fisheries Krishi Bhawan, New Delhi.
Negahdary, M., Hajihosseinlo, A. and Ajdary, M. 2013. PCR- SSCP variation of IGF1 and PIT1 genes and their association with estimated breeding values of growth traits in Makooei sheep. Genet. Res. Int, 2013, ID 272346.
Rasouli, S., Abdolmohammadi, A., Zebarjadi, A. and Mostafei, A. 2017. Evaluation of polymorphism in IGF-I and IGFBP-3genes and their relationship with twinning rate and growth traits in Markhoz goats. Ann. Anim. Sci., 17(1): 89-103.
Sambrook, J. and Russell, W.D. 2001. Molecular cloning. A laboratory manual. 3rd edn. Vol. 1 Cold spring harbor laboratory press, Cold Spring, New York, pp. 6.4-6.12.
Sankhyan, V., Thakur, Y.P and Dogra, P.K. 2019. Genetic polymorphism in IGF-1 gene in four sheep and goat breeds and its association with biometrical traits in migratory Gaddi goat breed of western Himalayan state of Himachal Pradesh, India. Indian J. Anim. Res., online first article DOI: 10.18805/ ijar.B-3795.
Thue, T.D. and Buchanan F.C. 2002. A new polymorphism in the cattle IGFBP3 gene. Anim. Genet., 33(3): 242.
Wang, P.Q., Deng, L.M., Zhang, B.Y. and Chu, M.X. 2011. Polymorphisms of the cocaine-amphetamine-regulated transcript (CART) gene and their association with reproductive traits in Chinese goats. Genet. Mol. Res., 10 : 731-738.
Wu-Jun, L., Guang-Xin, F., Yi, F., Ke-Chuan, T., Xi-Xia, H., Xin-Kui, Y, Mou, W., Hui, Y., Yong Zhen, H., Jing-Jing, H., Ya-Ping, X., Shi-Gang, Y. and Hong, G.H. 2010. The polymorphism of a mutation of IGF-1 gene on two goat breeds in China. J. Anim. Vet. Adv., 9(4): 790-794.
Zhang, C., Zhang, W., Luo, H., Yue, W., Gao, M. and Jha Z. 2008. A new single nucleotide polymorphism in the IGF-Igene and its association with growth traits in the Nanjiang Huang goat. Asian Austral. J. Anim. Sci., 21(8): 1073-1079.